Abstract
Introduction
Lymphomas of the central nervous system (CNS), including primary CNS lymphoma (PCNSL), have recently been focused on as promising targets for immune checkpoint blockade therapies. However, approximately one-third of patients with PCNSL are resistant to initial chemotherapies, and almost all patients will eventually develop recurrence or disease progression. During the clinical course, systemic extra-CNS lesions are observed in 3% of relapsed or refractory (r/r) PCNSL. To elucidate the mechanism of this uncommon recurrence suggestive of tumor oncogenic evolution and overcoming the immune privilege, we performed comprehensive genomic analysis comparing primary with systemic r/r PCNSL using next-generation sequencing approaches.
Methods
Patients at our institute with an episode of both PCNSL and extra-CNS relapse or disease progression with a pathological diagnosis of diffuse large B-cell lymphoma were screened. Genomic DNA was extracted from primary, recurrent, or refractory tumors, and corresponding germline samples. The samples were either formalin-fixed, paraffin-embedded (FFPE) or fresh frozen tissue if available. Clonal immunoglobulin gene rearrangements were detected using the IdentiClone™ IGH Gene Clonality Assay (Invivoscribe Technologies). DNA sequencing was performed on the Ion Torrent Proton™ platform after targeted amplification with the customized panel designed using the Ion AmpliSeq Designer™ tool (Life Technologies). This panel consists of whole exons of 64 genes associated with lymphoid malignancies, including those involved with the NFκB pathway, B-cell differentiation, epigenetic regulation of transcription, immune surveillance, and tumor suppression. Single nucleotide variations (SNVs), short insertions and deletions (InDels), and copy number variations were called using the Biomedical Genomics Workbench (Qiagen) after filtering and annotation using public variant databases such as dbSNP, COSMIC, and GRCh37 hg19. From these SNVs and InDels, somatic mutations were defined as non-synonymous, with a minimal coverage of 100, and with a frequency of subtracting germline reads from tumor reads of 10% or more. To confirm targeted DNA sequencing findings, we performed Sanger sequencing. To determine the cell-of-origin (COO), immunohistochemistry (IHC) using the Hans algorithm was performed. The expression of HLA class I, HLA class II, PD-L1, and PD-L2 was also assessed with IHC.
Results
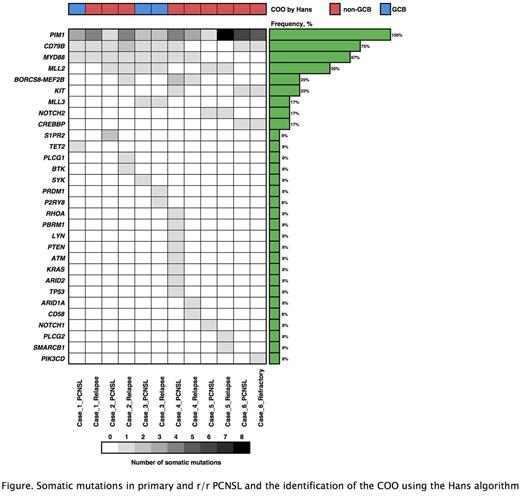
Six patients who were initially treated for PCNSL and who either relapsed (n = 5) or progressed (n = 1) were recruited. The relapse sites were the ileum (n = 2), testis with para-aortic lymph node (n = 1), intraabdominal lymph node (n = 1), and nasal sinuses (n = 1). The remaining refractory PCNSL was a patient who showed progression in multiple organs including the CNS with systemic lymph nodes. Only one relapsed patient was positive for HLA class I by IHC, with stronger staining seen at the relapsed site compared with the primary CNS site. Targeted DNA sequencing detected somatic PIM1 mutations in all primary and r/r tumors (Figure); nonsense mutations were detected in 50% of primary tumors and 67% of r/r ones. Compared to primary tumors, PIM1 was the most common gained mutation in r/r ones, and additionally gained somatic mutations such as CD79B, MLL2 and CD58 were observed. Partial loss of CIITA and the partial gain of NFκB pathway-related genes such as MALT1 and CARD11 were also detected. Somatic mutations were observed at a frequency of 1% in the germline sample of one patient (case 1 in Figure).
Conclusions
Our study suggested that the evolution of mutations in PCNSL enabled systemic disease progression with a breakthrough of immune privilege. These findings were characterized not only by an immunological overpowering but also by the dysregulation of B-cell proliferation signaling, suggesting that loss of function of PIM1 had occurred.
Toyoda: Chugai: Honoraria. Narita: Chugai, MSD, Eisai Co. Ltd., Nobelpharma Co.: Honoraria; Abbvie: Consultancy; Nihon Medi-Physics Co., Daiichi-Sankyo Co, Ono Pharmaceutical Co., AbbVie GK: Research Funding. Maruyama: Chugai, Kyowa Hakko Kirin, Ono, Celgene, Janssen, Eisai, Mundipharma, Takeda: Honoraria; Dai-ichi Sankyo, Chugai, Kyowa Hakko Kirin, Ono, Celgene, Janssen, GSK, Eisai, Mundipharma, Takeda, AbbVie, MSD, Sanofi, Pfizer, Otsuka, Novartis, Solasia, Zenyaku: Research Funding. Tobinai: Daiichi Sankyo Co., Ltd: Consultancy, Honoraria; Servier: Research Funding; Eisai: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Chugai: Honoraria, Research Funding; HUYA Bioscience: Honoraria; GlaxoSmithKline: Research Funding; Celgene: Consultancy, Honoraria, Research Funding; Kyowa Hakko Kirin: Honoraria, Research Funding; AbbVie: Research Funding; Takeda: Honoraria, Research Funding; Zenyaku Kogyo: Honoraria; Mundipharma: Honoraria, Research Funding; Ono Pharmaceutical: Honoraria, Research Funding. Kobayashi: Pfizer, Ohtsuka, Astellas, Ariad: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal